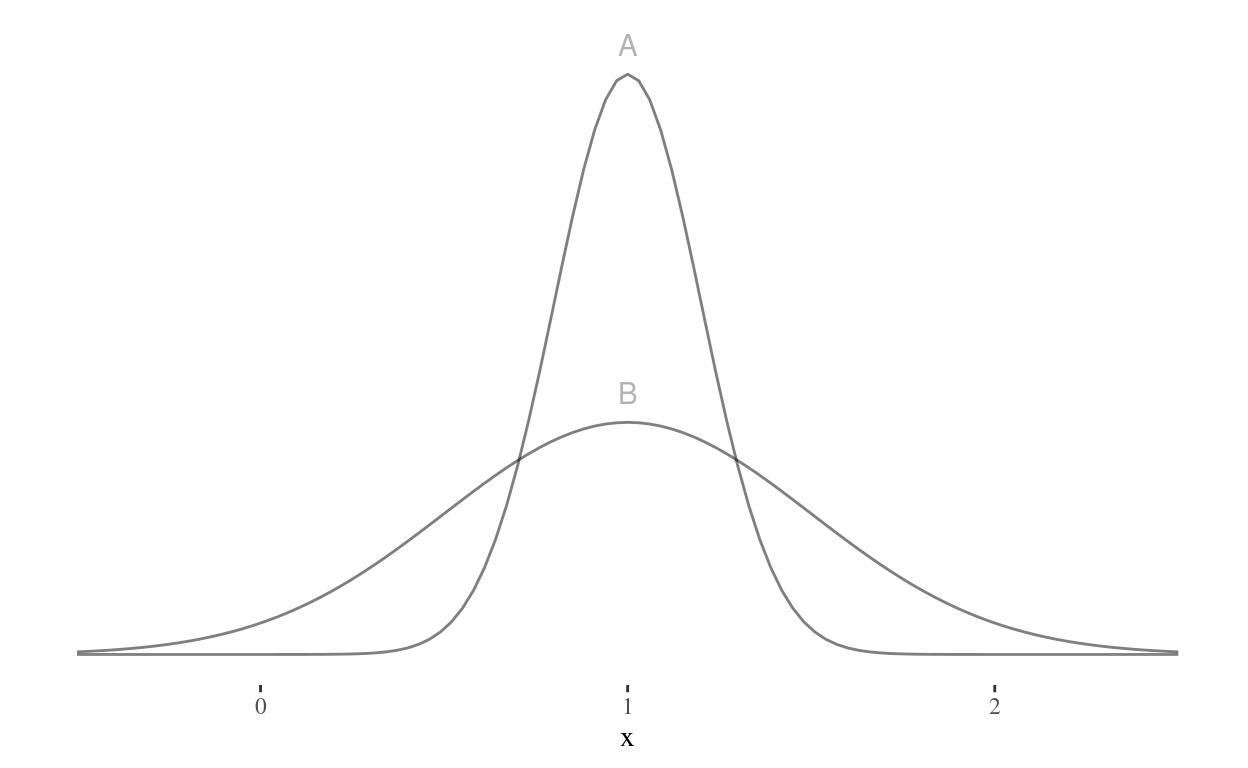
Suppose we have a study that has reported a p-value of for a t-test between groups A and B with the following means, standard deviations, and sample sizes.
dat %>%
gt()
| group | mean | sd | n |
|---|---|---|---|
| A | 1 | 0.2 | 35 |
| B | 1 | 0.5 | 50 |
ggplot() +
stat_function(
fun = dnorm,
args = list(mean = params$amu, sd = params$asigma),
alpha = 0.5
) +
stat_function(
fun = dnorm,
args = list(mean = params$bmu, sd = params$bsigma),
alpha = 0.5
) +
geom_text(
aes(x = x, y = y, label = label),
data = tibble(
x = c(params$amu, params$bmu),
y = dnorm(c(params$amu, params$bmu), mean = c(params$amu, params$bmu), sd = c(params$asigma, params$bsigma)) + 0.1,
label = c("A", "B")),
alpha = 0.3) +
theme_tufte() +
theme(
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
) +
xlim(min(c(params$amu, params$bmu)) - 3 * max(c(params$asigma, params$bsigma)),
max(c(params$amu, params$bmu)) + 3 * max(c(params$asigma, params$bsigma)))
Now we wish to write a simulation that draws a samples from each distribution of 35, 50, respectively, and computes a t-test, producing a p-value.
sample_a <- rnorm(params$an, mean = params$amu, sd = params$asigma)
sample_b <- rnorm(params$bn, mean = params$bmu, sd = params$bsigma)
sample_a %>% head()
[1] 1.0130998 1.0685929 0.6222748 0.9641765 1.0303178 1.1513003sample_b %>% head()
[1] 0.6239472 0.6695744 0.5656617 1.0382910 0.1608477 -0.2904730ttest <- t.test(sample_a, sample_b)
ttest
Welch Two Sample t-test
data: sample_a and sample_b
t = 1.0216, df = 63.378, p-value = 0.3109
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.08332685 0.25767415
sample estimates:
mean of x mean of y
0.9886130 0.9014394 # extract the p value
ttest$p.value
[1] 0.3108569Now let’s turn this into a fucntion.
ptrial <- function(
sim_id,
amu = params$amu,
bmu = params$bmu,
asigma = params$asigma,
bsigma = params$bsigma,
an = params$an,
bn = params$bn
){
# get samples
sample_a <- rnorm(n = an, mean = amu, sd = asigma)
sample_b <- rnorm(n = bn, mean = bmu, sd = bsigma)
# calculate t-test
ttest <- t.test(sample_a, sample_b)
# return pvalue
return(
tibble(
id = sim_id,
p_value = ttest$p.value
)
)
}
ptrial(1)
# A tibble: 1 × 2
id p_value
<dbl> <dbl>
1 1 0.374Now that we have a function, we can perform 120 trials.
| id | p_value |
|---|---|
| 1 | 0.03004456 |
| 2 | 0.99581508 |
| 3 | 0.06373585 |
| 4 | 0.36094549 |
| 5 | 0.98282944 |
| 6 | 0.88825267 |
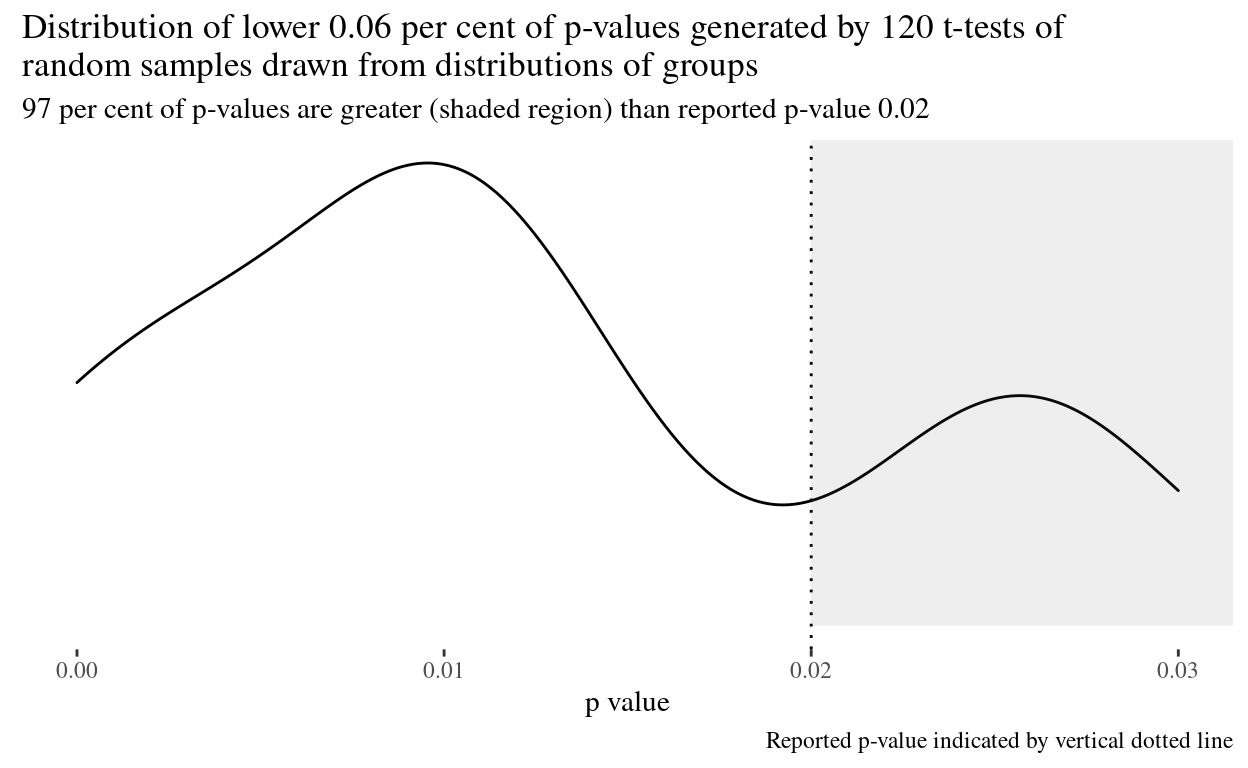
Now we plot the distribution of p-values.
reported_quant <- ecdf(trials$p_value)(params$reportedp)
shown_p <- params$reportedp + params$reportedp/2
shown_percent <- ecdf(trials$p_value)(shown_p) %>% round(2) * 2
trials %>%
ggplot(aes(x = p_value)) +
geom_density() +
theme_tufte() +
theme(
axis.line.y = element_blank(),
axis.ticks.y = element_blank(),
axis.text.y = element_blank(),
axis.title.y = element_blank()
) +
geom_vline(
xintercept = params$reportedp,
linetype = "dotted"
) +
annotate(
"rect",
xmin = params$reportedp,
xmax = Inf,
ymin = 0,
ymax = Inf,
alpha = 0.1
) +
labs(
title = str_wrap(glue("Distribution of lower {shown_percent} per cent of p-values generated by {params$trials} t-tests of random samples drawn from distributions of groups")),
subtitle = str_wrap(glue("{(1 - round(reported_quant, 2)) * 100} per cent of p-values are greater (shaded region) than reported p-value {params$reportedp}")),
x = "p value",
caption = "Reported p-value indicated by vertical dotted line"
) +
xlim(0, shown_p)