GEUS Application
Charles T. Gray
Ph.D. (Statistics) Towards a Measure of Codeproof: a toolchain walkthrough for computationally developing a statistical estimator
Why did I apply?
FAIR and Open Science
spunk package
Nutritional interventions for male infertility: a systematic review and meta-analysis
Open spunk data
Machine-readable data
library(spunk)
# data used in previous visualisation
spunk_dat %>%
filter(outcome == "morphology") %>%
sample_n(5) %>%
gt()| outcome | intervention | class | moderator | study | control | intervention_mean | intervention_sd | intervention_n | control_mean | control_sd | control_n |
|---|---|---|---|---|---|---|---|---|---|---|---|
| morphology | Folic acid | Vitamins | idiopathic | Silva 2013 | Placebo/no treatment | 23.91 | 3.68 | 23 | 24.23 | 3.06 | 26 |
| morphology | Selenium | Minerals | asthenoteratozoospermia | Safarinejad 2009b | Placebo/no treatment | 9.30 | 2.90 | 104 | 7.20 | 2.60 | 106 |
| morphology | Magnesium | Minerals | oligozoospermia | Zavaczki 2003 | Placebo/no treatment | 57.20 | 12.50 | 10 | 42.80 | 14.80 | 10 |
| morphology | Vitamin C + Vitamin E | Vitamins | oligoasthenoteratozoospermia | Greco 2005 | Placebo/no treatment | 8.00 | 7.10 | 32 | 11.60 | 7.80 | 32 |
| morphology | Lycopene | Antioxidants | oligoasthenoteratozoospermia | Nouri 2019 | Placebo/no treatment | 1.88 | 0.99 | 17 | 1.78 | 1.08 | 19 |
Code I’m proud of
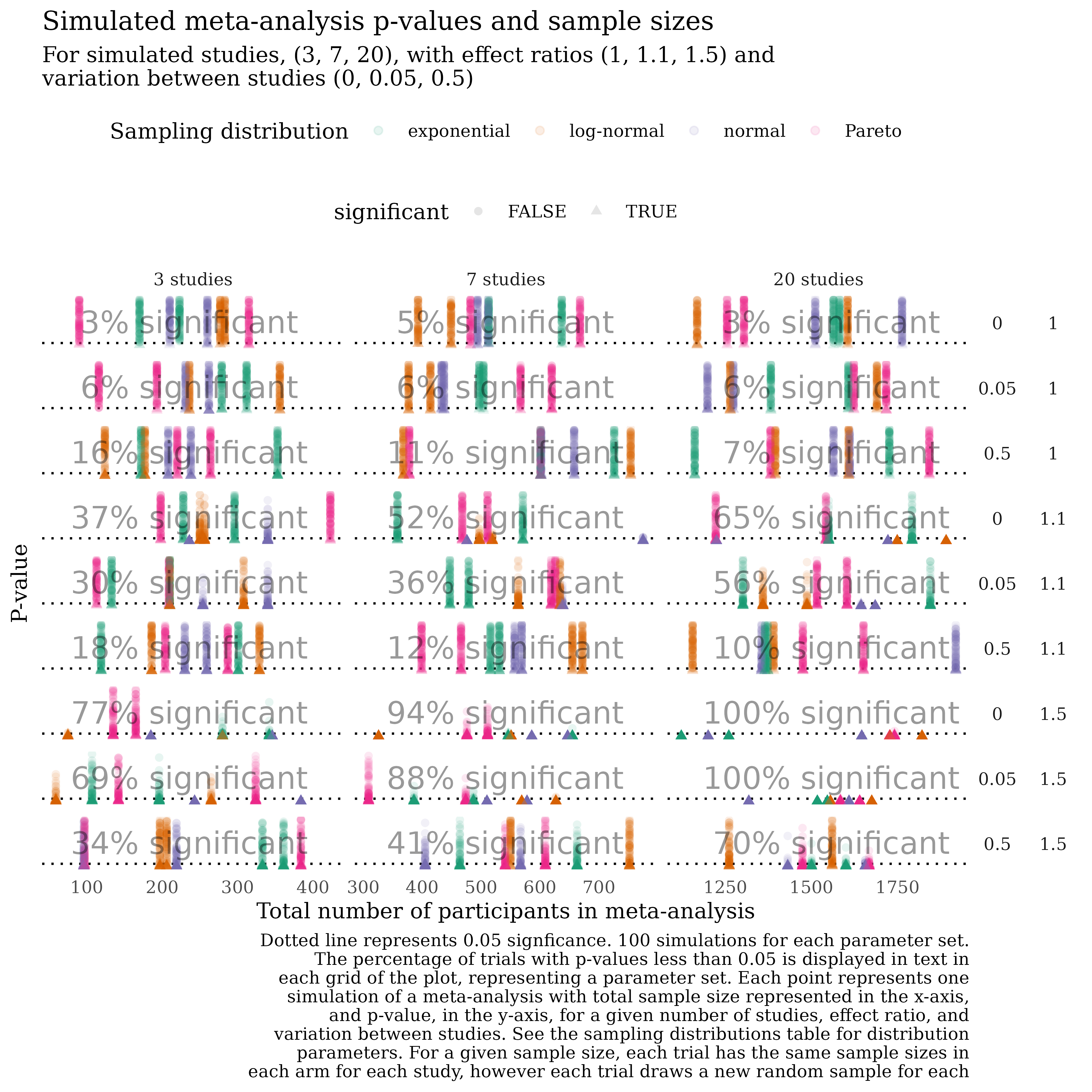
Simulating meta-analysis data
I have a question about simeta - I need to simulate meta-analyses with large, medium and small effect sizes and with high, medium and low heterogenity to illustrate the point about why vote-counting does not work (in all but the most extreme cases - large effect and no heterogenity). Can I do this using the code from simeta?
Uh, I don’t know
What I did
A brutally honest account.
- Procrastinated about responding
- Unchecked Bullet Journal task
- Set a meeting
- Opened Pandora’s Box (cloned repo)
- Check build
- Use-cases
- Meeting codejam
Packaged analysis: simeta
simeta

Math is in the manuscript, Statistics and Data Science, 2019
library(simeta)
# simulate dataset of 3 studies
example_meta <- sim_stats(
measure = "mean",
measure_spread = "sd",
n_df = sim_n(),
wide = TRUE, # metafor interoperability
rdist = "lnorm", # sampling distribution
par = list(shape = 0.25, scale = 1),
tau_sq = 0.4, # between-study error
effect_ratio = 1.2 # true effect
)
# output in html using quarto
example_meta %>% gt()| study | effect_c | effect_spread_c | n_c | effect_i | effect_spread_i | n_i |
|---|---|---|---|---|---|---|
| Elphir_1984 | 2.549209 | 2.727634 | 54 | 2.131478 | 1.944986 | 57 |
| Imrazôr_1992 | 2.605021 | 3.322262 | 86 | 3.071742 | 4.267402 | 88 |
| Yavanna_2015 | 1.749192 | 1.718549 | 30 | 3.973773 | 4.935866 | 27 |
What is a meta-analysis?
What is a meta-analysis?
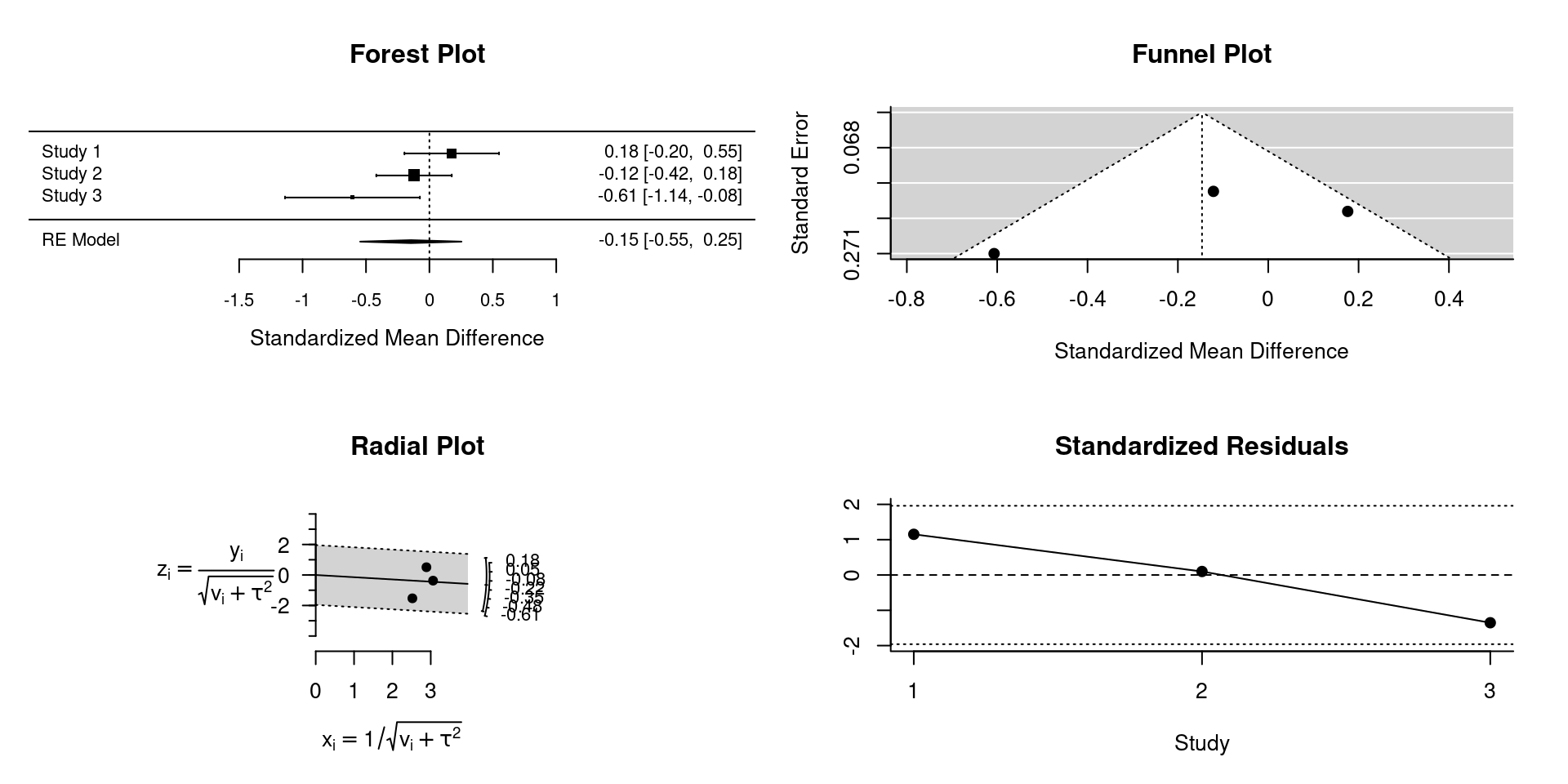
How was simeta implemented?
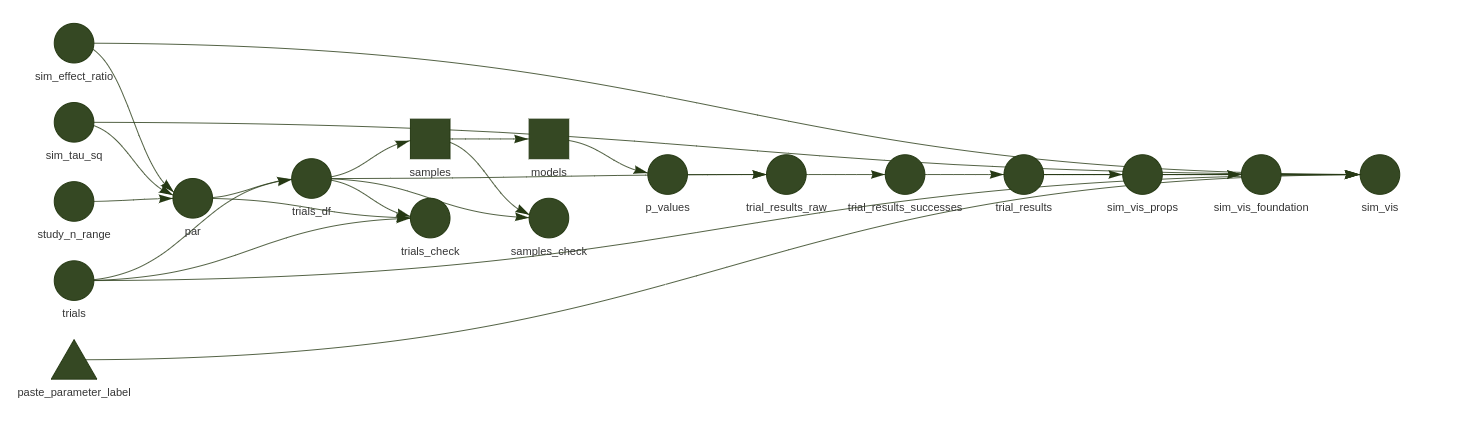
Pipeline to fit 25K meta-analysis models to simulated data, with parameters of interest extracted into a summarised in a ggplot visualisation.
- metafor models
- tidyverse tools
- roxygen documentation
- pkgdown site deployed on github pages
- targets simulation pipeline
What does it do?