cochrane
cochrane.Rmd
library(nmareporting)example data | Parkinson’s disease
# from data documentation
?parkinsonsmean off-time reduction in patients given dopamine agonists as adjunct therapy in Parkinson’s disease from 7 trials comparing four active drugs and placebo
parkinsons_dat %>%
# display top values
head(3)
#> studyn trtn y se n diff se_diff
#> 1 study_1 trt_1 -1.22 0.504 54 NA 0.504
#> 2 study_1 trt_3 -1.53 0.439 95 -0.31 0.668
#> 3 study_2 trt_1 -0.70 0.282 172 NA 0.282bayesian network meta-analysis model
\[ \left. \begin{array}{c r c l} \text{prior} & \boldsymbol d & \sim & \text{normal}(\boldsymbol d_0, \boldsymbol \Sigma_d)\\ \text{likelihood} & \boldsymbol y | \boldsymbol d & \sim & \text{normal}(\boldsymbol \delta, \boldsymbol V)\\ \text{fixed effects model} & \boldsymbol \delta &=& \boldsymbol{Xd} \end{array} \right\} \]
create network object
parkinsons_net <-
set_agd_contrast(
parkinsons,
study = studyn,
trt = trtn,
y = diff,
se = se_diff,
sample_size = n
)network meta-analysis model | prior
# prior
summary(normal(scale = 100))
#> A Normal prior distribution: location = 0, scale = 100.
#> 50% of the prior density lies between -67.45 and 67.45.
#> 95% of the prior density lies between -196 and 196.an example of an area I would like to learn more about; selection of priors.
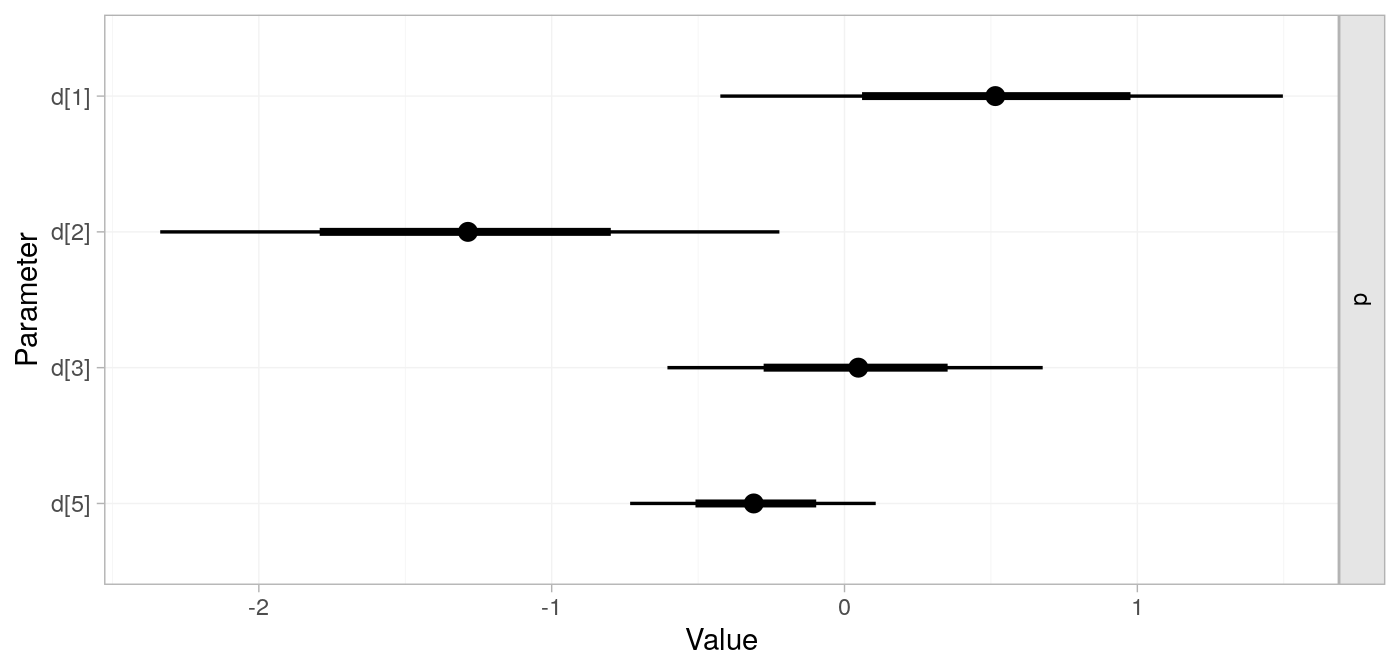
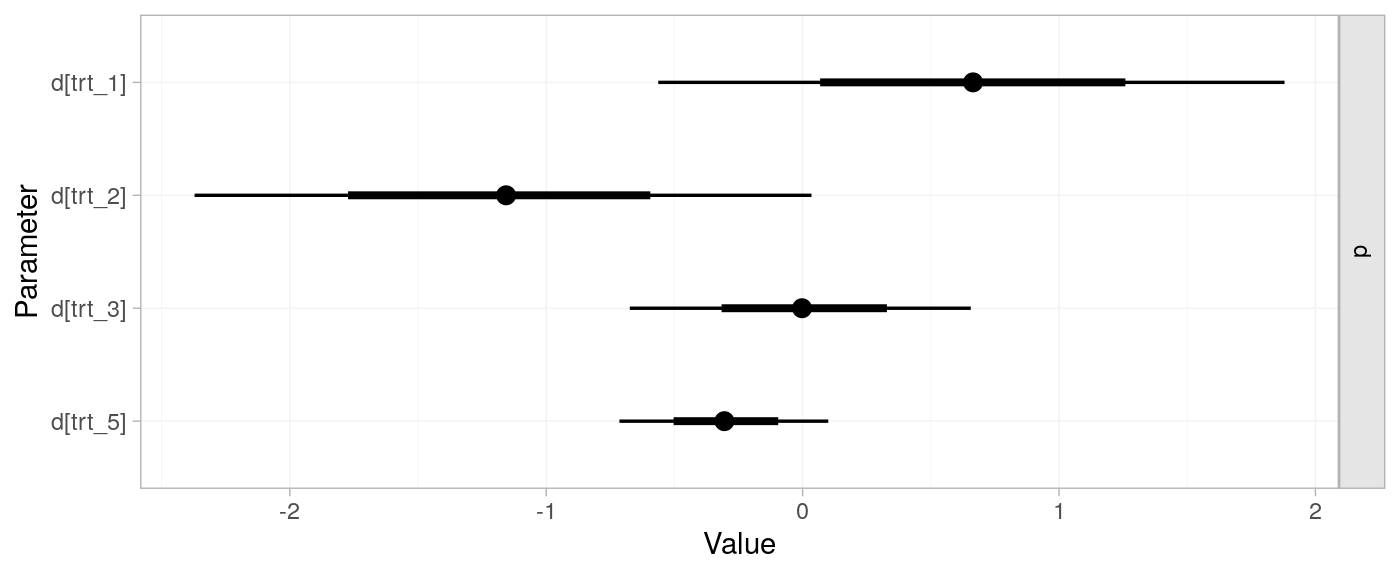
network meta-analysis model | ::nma
# fit model to network object
parkinsons_nma <-
nma(
# network object
parkinsons_net,
# fixed; for brevity w sensitivity
trt_effects = "fixed",
# set prior on treatment contrast with placebo
prior_trt = normal(scale = 100))
#> Note: Setting "4" as the network reference treatment.
#>
#> SAMPLING FOR MODEL 'normal' NOW (CHAIN 1).
#> Chain 1:
#> Chain 1: Gradient evaluation took 5.7e-05 seconds
#> Chain 1: 1000 transitions using 10 leapfrog steps per transition would take 0.57 seconds.
#> Chain 1: Adjust your expectations accordingly!
#> Chain 1:
#> Chain 1:
#> Chain 1: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 1: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 1: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 1: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 1: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 1: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 1: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 1: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 1: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 1: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 1: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 1: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 1:
#> Chain 1: Elapsed Time: 0.057321 seconds (Warm-up)
#> Chain 1: 0.050036 seconds (Sampling)
#> Chain 1: 0.107357 seconds (Total)
#> Chain 1:
#>
#> SAMPLING FOR MODEL 'normal' NOW (CHAIN 2).
#> Chain 2:
#> Chain 2: Gradient evaluation took 1.2e-05 seconds
#> Chain 2: 1000 transitions using 10 leapfrog steps per transition would take 0.12 seconds.
#> Chain 2: Adjust your expectations accordingly!
#> Chain 2:
#> Chain 2:
#> Chain 2: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 2: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 2: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 2: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 2: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 2: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 2: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 2: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 2: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 2: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 2: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 2: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 2:
#> Chain 2: Elapsed Time: 0.043281 seconds (Warm-up)
#> Chain 2: 0.04011 seconds (Sampling)
#> Chain 2: 0.083391 seconds (Total)
#> Chain 2:
#>
#> SAMPLING FOR MODEL 'normal' NOW (CHAIN 3).
#> Chain 3:
#> Chain 3: Gradient evaluation took 1e-05 seconds
#> Chain 3: 1000 transitions using 10 leapfrog steps per transition would take 0.1 seconds.
#> Chain 3: Adjust your expectations accordingly!
#> Chain 3:
#> Chain 3:
#> Chain 3: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 3: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 3: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 3: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 3: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 3: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 3: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 3: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 3: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 3: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 3: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 3: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 3:
#> Chain 3: Elapsed Time: 0.043368 seconds (Warm-up)
#> Chain 3: 0.046436 seconds (Sampling)
#> Chain 3: 0.089804 seconds (Total)
#> Chain 3:
#>
#> SAMPLING FOR MODEL 'normal' NOW (CHAIN 4).
#> Chain 4:
#> Chain 4: Gradient evaluation took 1.1e-05 seconds
#> Chain 4: 1000 transitions using 10 leapfrog steps per transition would take 0.11 seconds.
#> Chain 4: Adjust your expectations accordingly!
#> Chain 4:
#> Chain 4:
#> Chain 4: Iteration: 1 / 2000 [ 0%] (Warmup)
#> Chain 4: Iteration: 200 / 2000 [ 10%] (Warmup)
#> Chain 4: Iteration: 400 / 2000 [ 20%] (Warmup)
#> Chain 4: Iteration: 600 / 2000 [ 30%] (Warmup)
#> Chain 4: Iteration: 800 / 2000 [ 40%] (Warmup)
#> Chain 4: Iteration: 1000 / 2000 [ 50%] (Warmup)
#> Chain 4: Iteration: 1001 / 2000 [ 50%] (Sampling)
#> Chain 4: Iteration: 1200 / 2000 [ 60%] (Sampling)
#> Chain 4: Iteration: 1400 / 2000 [ 70%] (Sampling)
#> Chain 4: Iteration: 1600 / 2000 [ 80%] (Sampling)
#> Chain 4: Iteration: 1800 / 2000 [ 90%] (Sampling)
#> Chain 4: Iteration: 2000 / 2000 [100%] (Sampling)
#> Chain 4:
#> Chain 4: Elapsed Time: 0.040334 seconds (Warm-up)
#> Chain 4: 0.044476 seconds (Sampling)
#> Chain 4: 0.08481 seconds (Total)
#> Chain 4:cochrane reporting
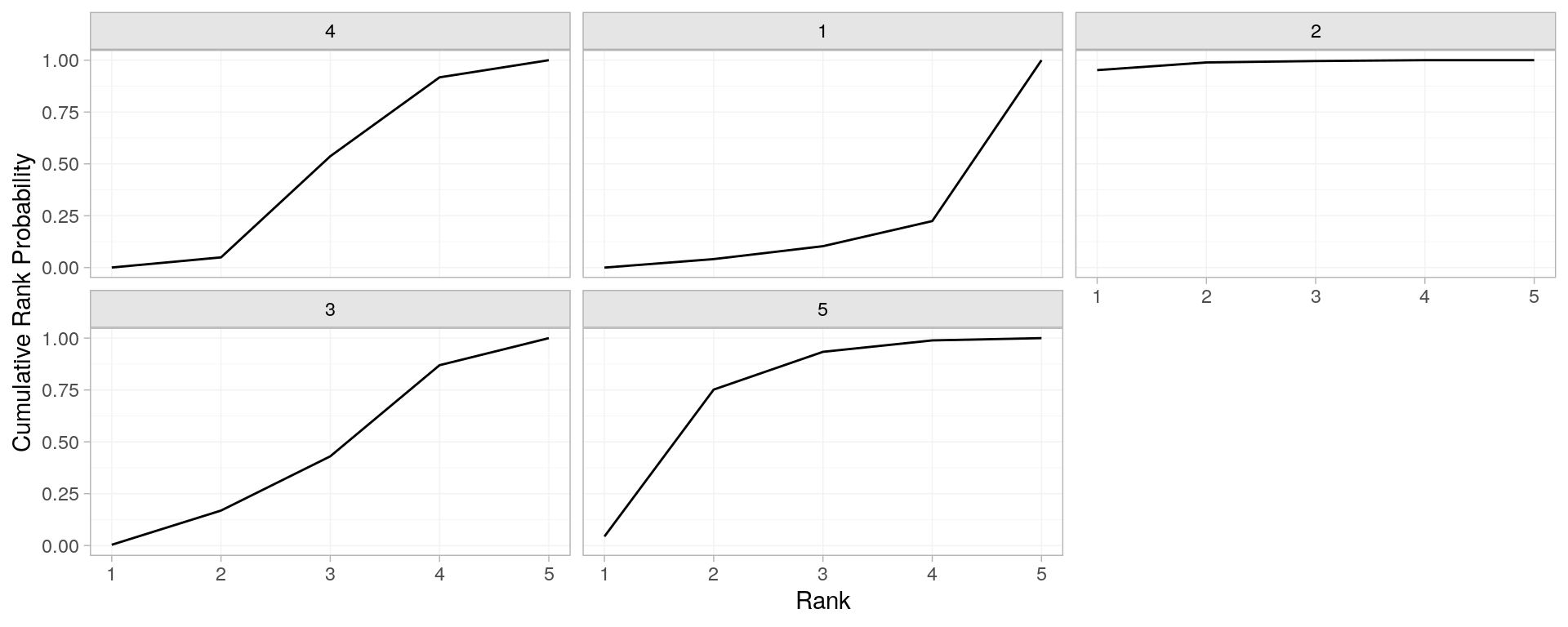
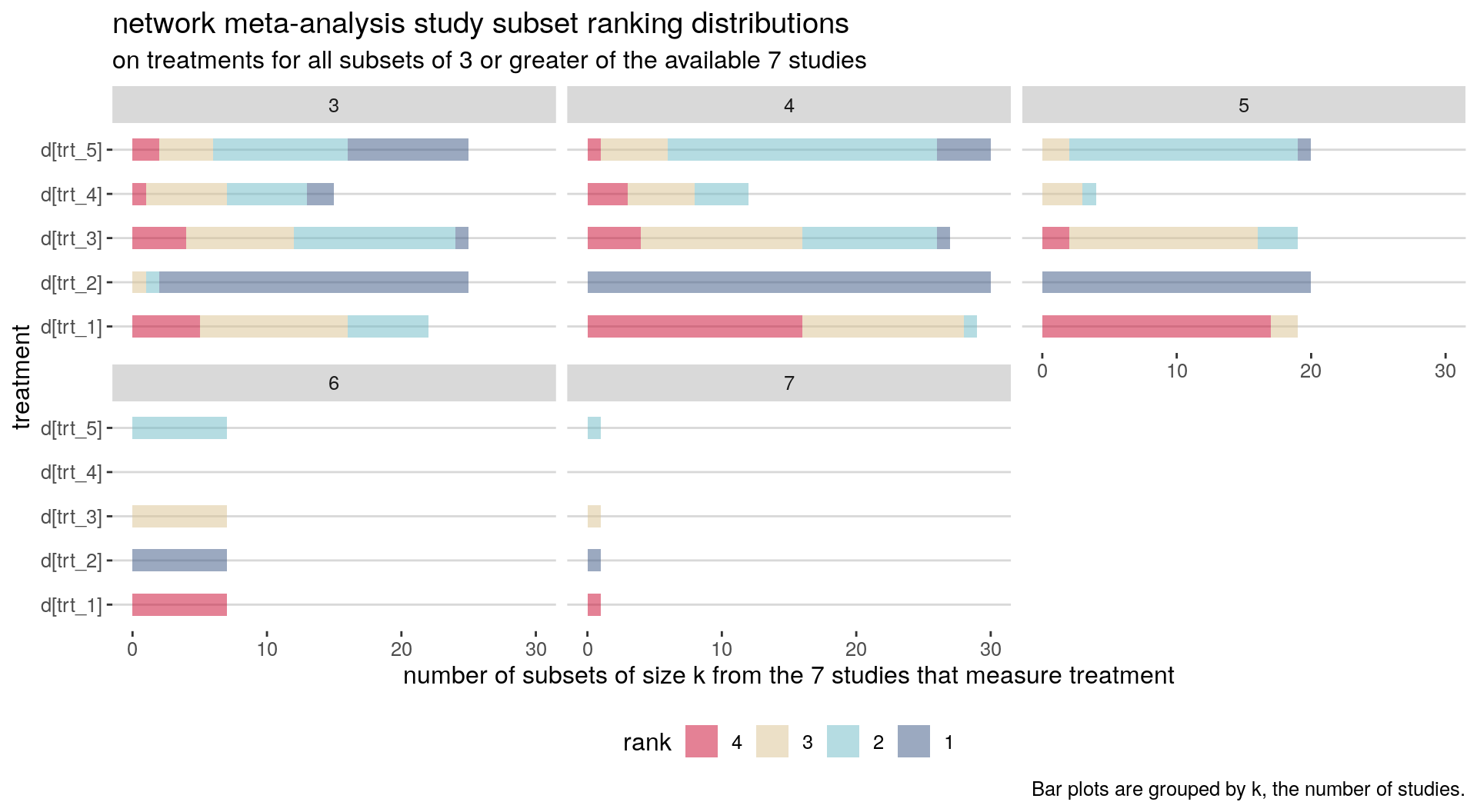
cochrane reporting | ranking
posterior_rank_probs(parkinsons_nma) %>% plot() 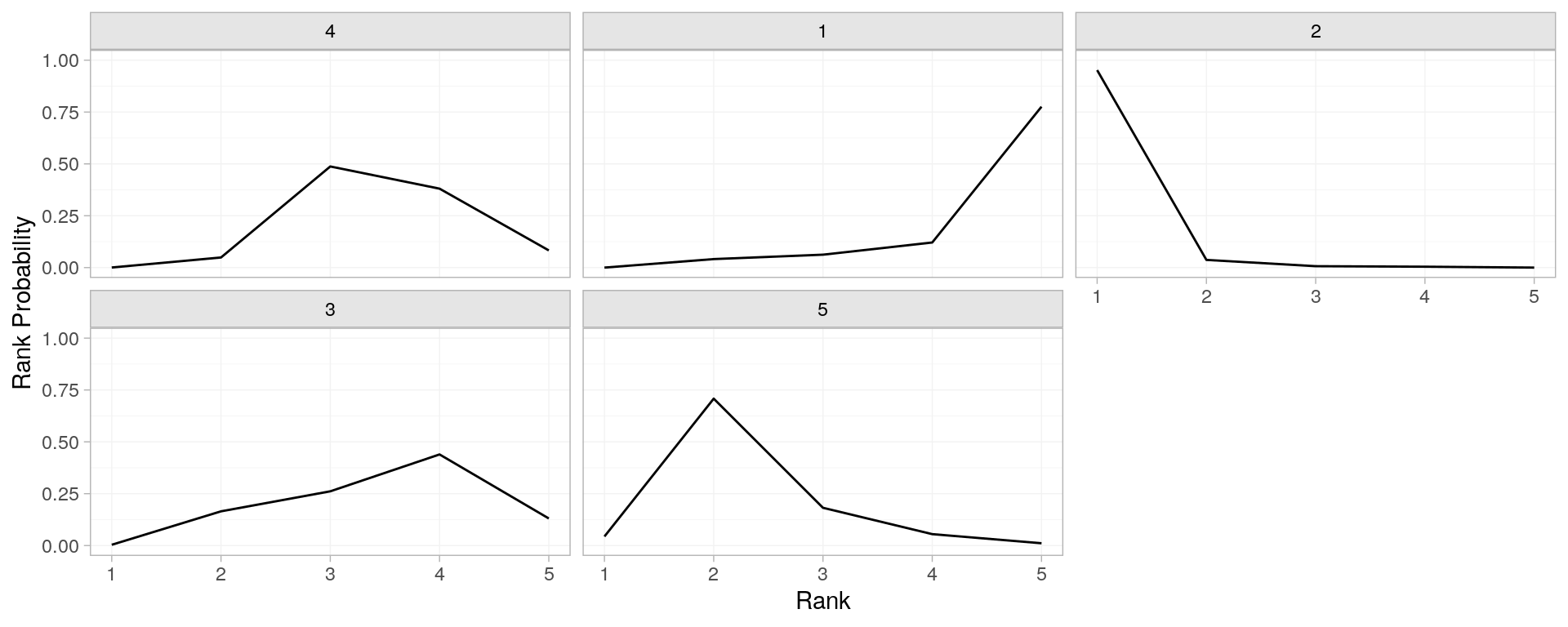
posterior_rank_probs(parkinsons_nma, cumulative = TRUE) %>% plot()